#Data Analysis 2: Cell Biology
#date 2024-30-01BIO00066I Workshop 2
Cell Biology Data Analysis Workshop 2
Workshop 1 for this module is here
The data you will work on in these workshops is the complete data set for the report. This is not example data.
1 Learning objectives
In this workshop, you will learn technical skills and data science concepts.
Technical skills
- RStudio skills to manage, view and analyse large data sets
- How to create plots in RStudio to assist with exploring and interpreting data
- How to make your data analysis reproducible and well-explained
Thinking like a data scientist
- How to interpret data with care
- How to integrate knowledge from laboratory work and computational analysis
The Module learning outcomes can be seen here.
2 Introduction
Workshops are not a test. It’s OK to make mistakes. It’s OK to need help. You should be familiar with independent study content before the workshop, but you don’t need to understand every detail. The workshop will hlpe you to build and consolidate your understanding. Tips:
- there are no stupid questions (ask us anything!)
- don’t worry about making mistakes (we all make mistakes)
- discuss code with your neighbours and with the staff
- outside of workshop times, google is a good resource
2.1 The biology
Today we will look at some data from mesenchymal stem cells (MSCs, sometimes called stromal cells). MCSs are are multipotent cells, than can differentiate into many kinds of mesenchymal cells, including bone, cartilage and fat cells. They are not pluripotent, in that they do have limits about how they differentiate.
MSCs are also:
- immuno-modulatory (they make cytokines)
- heterogeneous (they have subpopulations that are different from each other)
The MSCs we are looking at are from bone marrow femoral head tissues (from the hip bone). These MSCs were immortalised (by transforming them with a telomerase gene), and then cloned by limiting dilution. We will compare the data on cell shape and size from just two clones, both obtained from one person’s femoral head tissues. Each clone was derived from a single cell from the person. We call these two clones clone A and clone B. Today, we will use our data analysis skills to explore how these cell line differ. Remember, these two cell lines were both obtained from one person. You probably have these kinds of MSCs too.
2.2 Research questions
- What parameters do we have in the data obtained from the Livecyte machine/method?
- Which parameters differ between the clones?
- Which parameters are correlated?
2.3 The data
The data we have today are derived from live cell imagining of mesenchymal stem cells with a Livecyte microscope and the software it has. Each clones was cultured and the microscope captured cell shape and size information for thousands of cells from each clone (clone A and clone B). For each clone, we measured three biological replicates.
The Livecyte tracked each cell every 23 minutes, giving each data point a tracking ID (which tracks a unique cell ‘object’) and a lineage ID, which recalls the lineage of cells as they divide. For example, if cell 1, divided into cell 1A and cell 1B, they are all recorded by the Livecyte as being part of lineage ID 1.
This video shows what Livecyte can do (perhaps turn the sound down, or watch later).
A biological replicate repeats an experiment from different cell types, tissue types, or organisms to see if similar results can be observed.
Technical replicates are repeat measurements of the same biological material, which help to show how much variation comes from the equipment or different methods (rather from the biology).
Our replicates are almost biological replicates, because we grew the cells three times.
Data science can be challenging when you first start. But data science does have core concepts, like any other science.
- Consider the motivation or scientific question before drawing a plot
- Reflect on how the plot or analysis addresses the scientific question
- Use each plots to adapt, to inspire new research questions and new inquiries
- Ensure that your scripts are reproducible and clearly commented
- Consider what the results tell you about the biology
3 Exercises
As we run though the exercises, copy the code to your script. You can cop the code by clicking on the ‘clipboard’ symbol at the top tight of the grey code chunks.
#Comments In R scripts, lines that start with the hash symbol (#) are comments. Add comments so you can remember what this code does. Commenting code makes it readable, for you and anyone else. Good data science includes clearly commented code.
3.1 Getting started
- Start RStudio from the Start menu
- Open your RStudio project using the dropdown menu at the very top right of the RStudio window.
- Make a new script then save it with a sensible name that will help you to know later what is in this file.
BIO00066I-workshop2.Rwould work. - Add a comment to the script so you know what it is about, for example
- Clear all the previous data, and load
tidyversepackage by adding these lines to your script:
- Finally load another library that allows us to make multi-part plots (this can be useful sometimes). #we need this to make pretty plots with the ‘ggarrange’ package
The data you will work on in these workshops is the complete data set for the report. This is not example data.
- Make sure all these lines of code are in your script, with comments.
- Save the script.
3.2 Loading the data
In workshop 1, we showed you how to import data from files. Today, we load a tab-separated value (TSV) file from a website:
cells <-read_tsv(url("https://djeffares.github.io/BIO66I/all-cell-data-FFT.filtered.2024-02-22.tsv"),
col_types = cols(
clone = col_factor(),
replicate = col_factor(),
tracking.id=col_factor(),
lineage.id=col_factor()
)
)We use the read_tsv function to read the tab-separated value file. Clicking on the link in the read_tsv takes you to a website about this function. All the code chunks in these workshops have links like this. The clone = col_factor(),replicate = col_factor() etc let’s R know that we want these columns to be factors, rather than numeric values. Factors are used to represent categorical data.
3.3 Exploring the data
It is important to know what data you have. How many rows and columns etc. There are many ways to do this in R. Here are some of our favourites. Copy and past these into your R script, and try them out.
You may have noticed that we have two clones (cloneA and cloneB). For each clone, we have three replicates. For each replicate, we have many readings of cell widths, cell volume, cell sphericity, and so on.
Why not take a note of this, and use it all the time?
3.4 Cleaning up
When you ran names(cells) you will have seen many metrics from the Livecyte software. We don’t need all these, and the movement metrics them are unreliable, so let’s remove them. It is simple to remove columns of data, using the select command, like so:
#see what we have
names(cells)
#remove all the movement metrics (which are not reliable)
cells <- select(cells,
-position.x,
-position.y,
-pixel.position.x,
-pixel.position.y,
-displacement,
-instantaneous.velocity,
-instantaneous.velocity.x,
-instantaneous.velocity.y,
-track.length
)
#check that our data is simpler
names(cells)In the select function, using the minus sign - before a column name removes it. We can also also use select to define which columns to keep, by omitting the -. For example cells <- select(cells, clone, replicate, volume) will only keep the columns we listed.
3.5 Save your work regularly
Now save your data and save your script. This command will save all the variables you have loaded, or created so far:
#save all my stuff
save.image("BIO00066I-workshop2.Rda")You can load all the data again with:
#load all my stuff from last time
load("BIO00066I-workshop2.Rda")3.6 Summarise with dplyr
The view(cells) command shows that we have many cell shape metrics. We saw a summary of our cell shape metrics with the summary(cells) command above.
We can do even better, with tools from the dplyr package (part of the tidyverse). These allow us to make summaries of this data quickly. These methods will work for any data frames of any kind of data.
dplyr is like a set of pliers, helping us to ‘bend’ or ‘reshape’ our data.
Here is the command. I will explain it below.
3.6.1 What this code does
The
cells |>part takes the data from thecellsdata frame, and ‘pipes’ it into thegroup_byfunction. The|>symbol as means put this data into the next bit.group_by(clone, replicate)means that make groups of data, according to which clone they are, and which replicate culture they were from.summarisecalculates some summaries of all the data rows (for each clone and replicate). The partvolume=median(volume)creates a header calledvolumeand fills this with the median cell volume for each clone and replicateRight at the top,
summary.table <-stores the results of all the piping in an object calledsummary.table.
Have a look at the information we generated with:
view(summary.table)- What does this table tell you about clone A and clone B?
- What does this tell you about mesenchymal stem cells?
3.7 Making plots
It looks like many of the cell shape metrics from above might differ between clones. So let’s look deeper, starting with cell width. We will make a box and whisker plot. We use a small ‘trick’ here: by including fill=replicate we force R to make different plots for each replicate.
ggplot(cells,aes(x=clone,y=width,fill=replicate))+
geom_boxplot()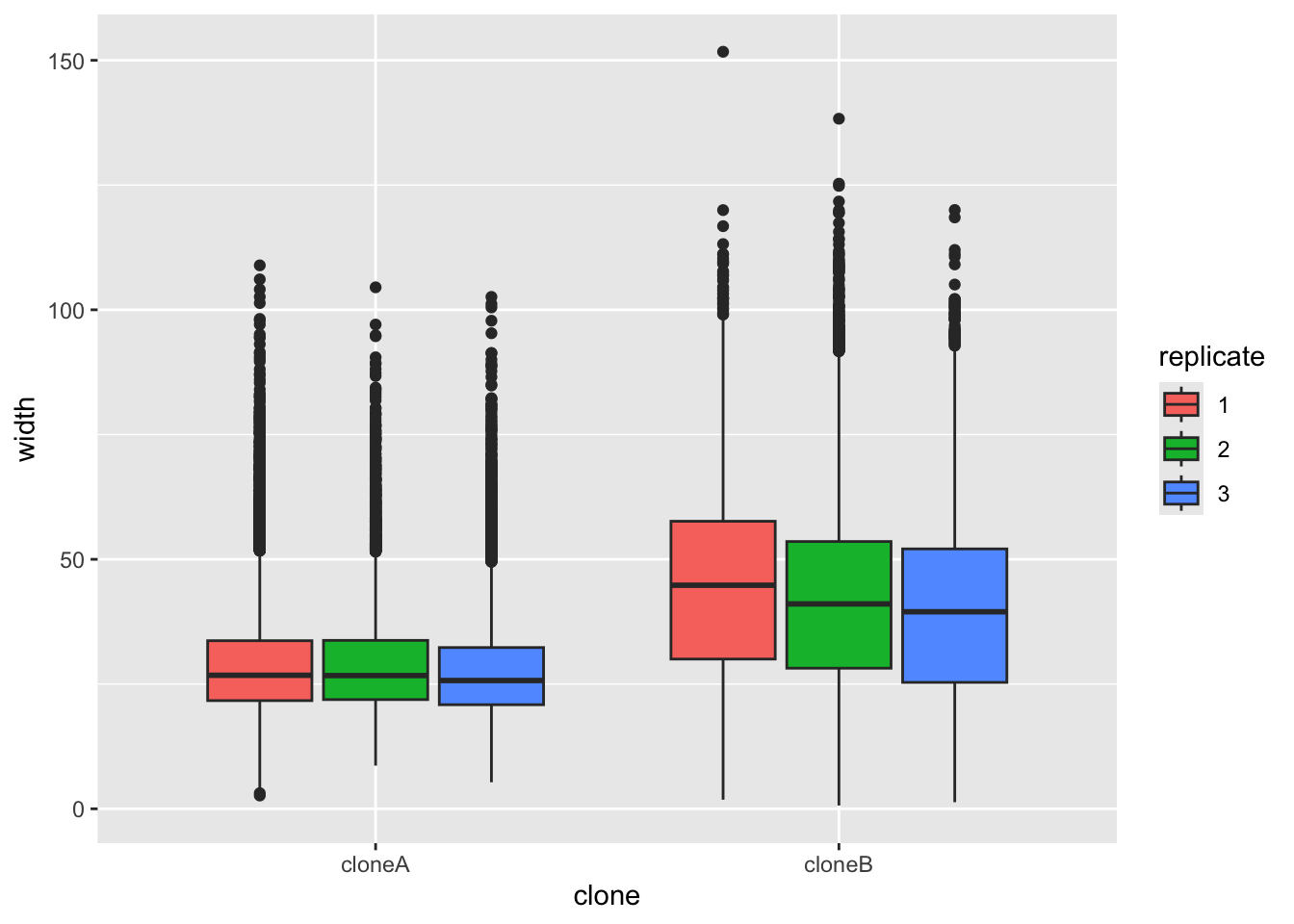
No matter what shape plot you want, ggplot uses the same syntax.
- Start by telling R what data you want to use like this:
ggplot(some_data_frame,aes(x=x_axis_data, y=y_axis_data) +
In this line, x_axis_data and y_axis_data are columns of some_data_frame.
- Then define what the shape plot you want (the ‘geometry’):
geom_boxplot(),geom_histogram()etc. - Add extra things to customise your plot, eg:
xlab("my x axis label")
If you are unsure what to do, it’s OK to ask! Googling “ggpplot how to make a boxplot” (or any other plot) will help.
It does look like clone A and clone B differ consistently in width, with all repeats. To prove this, we would like to do a statistical test. A Student’s t-Test t.test would work. But t-Test’s assume that the data are normally distributed (like a bell curve).
We can test this approximately by plotting data, as below. It doesn’t look perfectly ‘bell shaped’, so let’s play is safe and use a nonparametric test.
ggplot(cells, aes(x = width)) +
geom_density()The nonparametric equivalent of a t-Test is a Wilcoxon rank sum test. There are two ways to run this test in R. We can do wilcox.test(vectorA, vectorB), where vectorA and vectorB contain the numeric values we want to test.
But does does not suit tidyverse data frames very well. So we will use the wilcox.test(numeric_values ~ category_name, data = some_data_frame) method, like so:
#Wilcoxon rank sum test
#To test if cloneA and cloneB have statistically different widths
wilcox.test(width ~ clone, data = cells)
Wilcoxon rank sum test with continuity correction
data: width by clone
W = 657964994, p-value < 2.2e-16
alternative hypothesis: true location shift is not equal to 0Because the p-value is very low (< 2.2e-16), this means the null hypothesis (that both sets of numbers come from the same population) is very unlikely to be true.
Parametric tests are based on assumptions about the distribution of the real data. Nonparametric statistics are not based on these assumptions. So they are safer. Many biological metrics need nonparametric tests.
3.7.1 An easier way
Fortunately, you can add the results of nonparametric tests to ggplot, by adding stat_compare_means() to our plot. Note that below we do not force R to split up the replicates as we did above.
ggplot(cells,aes(x=clone,y=width))+
geom_boxplot()+
stat_compare_means()4 Next step: modify the code
To increase your understanding, run through all the plots in section 3.5 with some other cell shape or size metric. To start, find what metrics are present with:
names(cells)We suggest using one of these, in place of width:
- volume
- mean.thickness
- radius
- area
- sphericity
- length
- dry.mass
5 Analysis with small data
So far, we have used all the rows in our data frame. But perhaps this isn’t wise, because the Livecyte tracking followed each cell over time. So each measurement of cell shape is not a completely independent measurement. It is more of a technical replicate than a biological replicate.
It is also valuable to learn how to make plots and analyse small data sets, as well as large ones. So not we will create a very small subset of the data, and investigate this.
First, read in the small data set we prepared, and have a quick look, to check what we have.
#read in some data from a website
small.tracking.data <-read_tsv(url("https://djeffares.github.io/BIO66I/trackingid.small.data.tsv"),
col_types = cols(
clone = col_factor(),
replicate = col_factor(),
tracking.id=col_factor(),
lineage.id=col_factor()
)
)
#check what we have
glimpse(small.tracking.data)
summary(small.tracking.data)5.1 Plotting a small data set
Previously we used geom_boxplot() to compare clone A and clone B. Box and whisker plots (geom_boxplot) and violin plots (geom_violin, which we describe below) are good ways of displaying large amounts of data, but they can be misleading for small data sets. For small data sets it is better to simpler plot all the data points, so the audience has all the information. Let’s compare boxplot and a ggplot method to plot all the data (geom_jitter).
ggplot(small.tracking.data, aes(x=clone, y=width))+
geom_boxplot(fill=NA)+
geom_jitter(width = 0.2,size=3,pch=1)+
theme_minimal()
names(small.tracking.data)Try two alternations of this plot.
Remove the
geom_boxplot(fill=NA)+line. Does it display the data more clearly?Replace width in
y=widthwith some other cell shape/size parameter. Usingnames(small.tracking.data)will show what metrics we have.
5.2 Statistical tests with small data and large sets
We use statistical tests like the Wilcoxon rank sum test (wilcox.test) to estimate how likely to be drawn from the same ‘population’ (or distribution). If the P-value is very low (eg: one chance in 10,000 or 0.0001), then we can conclude that the two data sets are indeed different. But the power of such a test depends on the number of data points.
We can prove that to ourselves by comparing a Wilcox test results from the small.tracking.data and the much larger cells data set:
#test with small.tracking.data
wilcox.test(width ~ clone, data = small.tracking.data)
Wilcoxon rank sum exact test
data: width by clone
W = 75, p-value = 0.1261
alternative hypothesis: true location shift is not equal to 0#test with 'cells' data frame - a *much* larger data set
wilcox.test(width ~ clone, data = cells)
Wilcoxon rank sum test with continuity correction
data: width by clone
W = 657964994, p-value < 2.2e-16
alternative hypothesis: true location shift is not equal to 0P-values are not the biology. Use them with caution, and your own judgment. Sometimes very small differences, or very weak correlations can be statistically significant, but have little relevance for the biology.
5.3 Comparing plots side by side
Now, lets look plotting the small and large data sets side by side. Sometimes, a plot can give us a better sense of the data that a P-value.
#plot with the large data
#storing the plot in an object called large.data.plot
large.data.plot <- ggplot(cells, aes(x=clone, y=width))+
geom_boxplot()+
geom_jitter(width = 0.2,size=3,pch=1)+
stat_compare_means()+
ylim(0,200)+
ggtitle("large data")
#plot with the small data
#storing the plot in an object called small.data.plot
small.data.plot <- ggplot(small.tracking.data, aes(x=clone, y=width))+
geom_boxplot()+
geom_jitter(width = 0.2,size=3,pch=1)+
stat_compare_means()+
ylim(0,200)+
ggtitle("small data")
#create a two panel plot, with large.data.plot and small.data.plot
ggarrange(large.data.plot,small.data.plot)The geom_jitter with the large data set looks terrible! This is not a good way to show this data. So remove this part of the plot, and try again. You may also want ot try using geom_violin instead of geom_boxplot.
You will also notice that we set the same y axis range for both plots, with ylim(0,200). We did this so we can compare the plots side by side.
6 Reflection
- After loading the data, what did we do first?
- What did making the plots do that summary statistics (liek average, or median) cannot do?
- What did we learn about the two clones?
- How do we plot and consider P-values for small and large data sets?
7 The end
This is the end of the workshop for today. We provide some consolidation exercises, extra material and advice about the R Studio project below.
8 After the workshop
Look at your script again. Add some comments, so that the next time you look at it, it will make more sense to you. Also, have a go at the consolidation exercises below.
8.1 Consolidation exercises
8.1.1 Violin plots geom_violin
- We used
geom_boxplot()to show the differences between categories (clone A and clone B). Trygeom_violininstead. Which one do you prefer?
ggplot(cells,aes(x=clone,y=width,fill=replicate))+
geom_violin()- Now adjust the title with
ggtitle(label), and the axis labels withxlab(label)andylab(label), and the theme (the general style of the plot), withtheme_classic(). There are many themes for ggplot, which you can see here. Use whatever theme you feel makes the data clear and simple.
ggplot(cells,aes(x=clone,y=length,fill=replicate))+
geom_violin()+
ggtitle("put your title here!")+
xlab("my X axis label")+
ylab("my Y axis label")+
theme_classic()8.1.2 Making the small data set
This is how we made the small data set trackingid.small.data. Reading through and/or trying out this code will enhance your skills.
Use the dplyr tools group_by and summarise to calculate the average of all the cell shape measurements for each cell. Each cell is marked by the Livecyte software with a tracking id (tracking.id), so we group cells by this. The code group_by(clone, replicate, tracking.id) will group each measurement by the clone, replicate and tracking id, then the summarise part calculates the mean values for all our metrics:
#take the mean of each unique tracking id
trackingid.summary.table<-cells |>
group_by(clone, replicate, tracking.id) |>
summarise(
volume=median(volume),
mean.thickness=median(mean.thickness),
radius=median(radius),
area=median(area),
sphericity=median(sphericity),
length=median(length),
width=median(width),
dry.mass=median(dry.mass),
length.to.width=median(length.to.width)
)`summarise()` has grouped output by 'clone', 'replicate'. You can override
using the `.groups` argument.Then we use the dplyr function sample_n, which samples a number of rows from a data frame at random. If the data frame is grouped (which our data is),the number we set applies to each group:
We write the output to a file:
#output trackingid.small.data as a tab-separated value (tsv) file
write_tsv(trackingid.small.data, file ="/Users/dj757/gd/modules/BIO66I/data/trackingid.small.data.tsv")Look at the trackingid.small.data data frame, so we know what we have:
8.2 Planning for your report
For this module, 30% of the grade is for an R Studio project. The script that you are creating now is the start of this. Here is some information that will help you to prepare to submit your project.
Create a logical folder structure for your analysis. Your submission should include the script you used for your work, any accessory functions and the data itself. Your script should be well-commented, well-organised and follow good practice in use of spacing, indentation and variable naming. It should include all the code required to reproduce data import and formatting as well as the summary information, analyses and figures in your report.